|
|
 |
 |

|

|

|
|
Drug Discovery
The Iterative Drug Discovery (IDD) Platform represents a paradigm shift in drug discovery methodology. Based on gene expression methods and knowledge about genome pathways and the complex interactions among genes, it leverages our microarray and computational expertise to attack the target selection efficiency problem in the specific field of expression control, and rapidly:
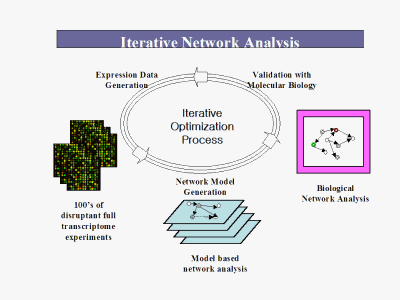
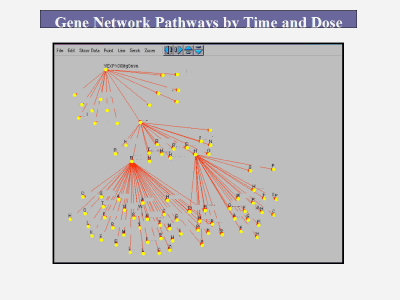
The IDD Platform includes: High Value Proprietary Transcriptome Data: We begin by constructing large scale, highly accurate, full genome transcriptome libraries on our proprietary microarrays. These libraries consist of hundreds of results of single gene deletions. Thus, we have data on the effect of each individual genes presence or absence on the expression of all the other genes in the genome. Proprietary Transcriptome Network Algorithms: We then use our proprietary, patent pending algorithms to take the transcriptome data and use it to reverse engineer the transcription regulatory relationships among all the genes in the genome. Targeting: The networks are then used as the skeleton framework for target selection and optimization. From this information, high potential targets are assessed with molecular biological validation for phenotype. Because it elucidates the inner, "black box" workings of intermediate gene expression regulation events (i.e. gene to gene interactions and cascades that produce the phenotype) between lead molecule/biological target and phenotype that are ignored by current methods, it facilitates wiser target selection and decreases the risk of drug failure from poor efficacy or toxicology problems. Importantly, it can be extended to work in reverse from a given phenotype/outcome to identify molecules such as receptors that will generate the desired phenotype.
Once the contents of the network "black box" are known, the network analysis becomes the hub of a reliable target generation machine that can identify high-worth, low-risk targets from three different starting points: Biological: Knowledge of known receptors, pathways and molecules along with the added information of the network allows our scientists to find novel, high-worth targets from present knowledge. An example of this is the identification of an orphan nuclear receptor "master switch" for DNA repair mechanisms that was discovered using the platform. Small Molecule: Using a patent pending method to combine our model transcriptome data with transcriptome data from drug response experiments, we can directly identify the regulatory pathways of action of a given lead molecule and then use this information to identify better alternative targets. Clinical Phenotype: Similarly to "Small Molecule" above, transcriptome data from patient and normal samples can be combined with the platform to generate selected regulatory molecules of direct clinical relevance. 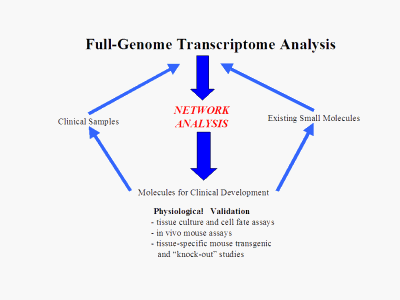
|